Emma Devereux
Emma Devereux - Monitoring wildlife health in the Kruger National Park: Can gut microbiome provide a noninvasive indicator of health?
Emma Devereux, SVM, Class of 2021, University of California, Davis
Drs. Patricia Conrad, Terra Kelly, & Brian Bird (UCD); Dr. Brianna Beechler (Oregon State University)
Hypothesis: Assessing wildlife health often relies on capture and sedation of animals. This process is expensive and often infeasible for some rare or elusive species. Handling also exposes animals to undue risks such as fractures, capture myopathy and other morbidity or mortality causing events. These challenges underscore the need for a noninvasive strategy for monitoring wildlife health. The use of gastrointestinal microbiome for detecting animal health is a potentially viable strategy for addressing this need for several reasons: (1) the information required to characterize an animal's gut microbiome can be gathered from fresh feces, which can be obtained easily from most species without handling or exposing the animal to unnecessary risks; (2) recent metagenomic sequencing innovations have driven down costs of data generation and enabled the characterization of microbiome composition based on a single, taxonomically diagnostic gene (e.g. 16s), making this approach both cost-effective and feasible; and (3) established links between disease status and gastrointestinal microbiome implicate gut microbiota as a viable potential indicator of animal health (1, 2, 3). This research seeks to correlate gut microbial communities, as assessed by collection and analysis of fresh feces, with routine health measures to support the use of GI microbiome as an alternative indicator of animal health. Our hypothesis was that the diversity, richness and structure of gastrointestinal microbial communities will be correlated with traditional measures of animal health, both within single taxa and across multiple taxa of herbivorous host species.
Research accomplished: Feces and blood were collected from all herbivorous animals darted and/or captured by the Veterinary Wildlife Services (VWS) team in the Kruger National Park (KNP) between June 1st and July 18th, 2018. Observations were limited to herbivores as diet is known to be a strong predictor of GI microbial community structure (4). Body condition scores (1-5), skin condition (i.e. presence of mange), environmental conditions and pregnancy/lactation status were recorded for all captured individuals. Hematocrit and total protein were evaluated from blood samples, and blood smears were prepared and analyzed under a microscope at the VWS laboratory – total white blood cell count and cell differentials were calculated and recorded. DNA was extracted from feces using the MoBio powersoil DNA extraction kit and transported back to Oregon State University for gut microbiome sequencing in the Sharpton Lab. Due to unexpected delays in the OSU lab, sample sequencing will commence in September. In the meantime, hematological data were analyzed using GraphPad Prism 7 software.
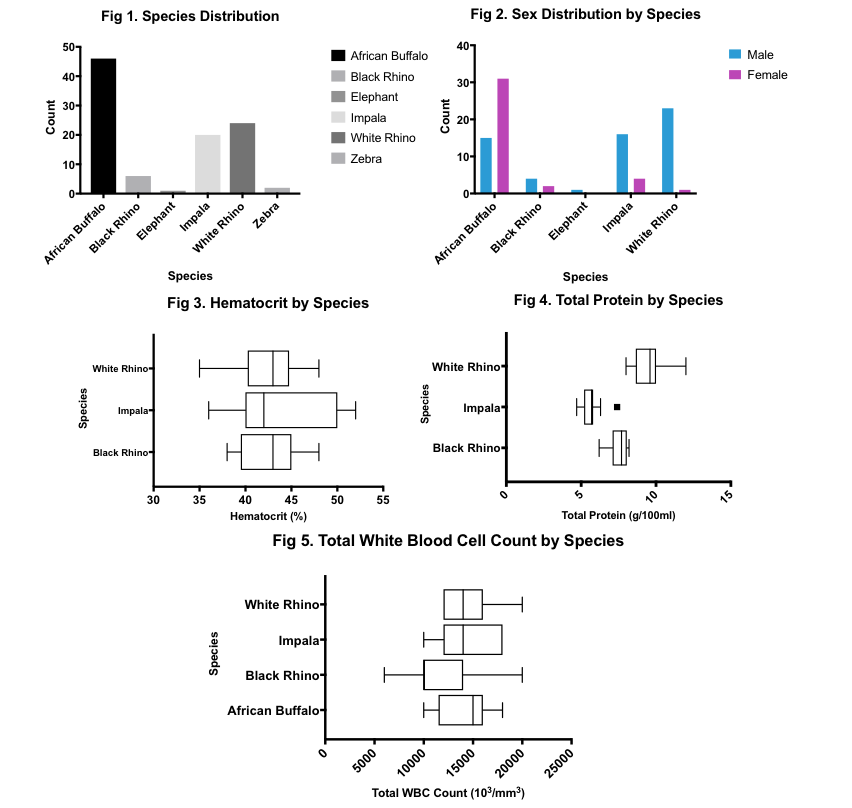
Results: Samples were collected from 99 individuals in the Kruger National Park between June 1st and July 18th, 2018. Sampled individuals belong to six herbivorous species (Fig 1), including African Buffalo (n=46), Black Rhino (n=6), Elephant (n=1), Impala (n=20), White Rhino (n=24), and Zebra (n=2). The majority of sampled individuals are male for all species, except for African Buffalo (Fig 2). Hematocrit values were similar across all species for which n>5 blood samples (p=0.5885; Black Rhino=42.67%; Impala=43.91%; White Rhino=42.46%) (Fig 3). Total protein varied between species for which n>5 blood samples (p<0.0001; Black Rhino=7.533g/dL; Impala=5.645g/dL; White Rhino=9.492g/dL) (Fig 4). Similarly, total white blood cell count varied between species for which n>5 blood smears (p=0.0002; African Buffalo=14,200; Black Rhino=11,333; Impala=14,545; White Rhino=14,400) (Fig 5). Lymphocytes were the dominant cell type in African Buffalo and Impala, while neutrophils dominated in both Black and White Rhino. Body condition score did not prove to be a useful indicator of health in this context due to minimal spread and lack of variance in the data. However, hematocrit has been shown to significantly and positively correlate with body condition score in mammals and will serve as a proxy for body condition score for the purposes of future analyses (5). Although these parameters have been previously explored in the literature, understanding the spread of hematological values in our particular sample population was a critical first step towards correlating the diversity, richness and structure of gut microbial communities with traditional, blood-based measures of health in herbivorous host species.
Appendix.
Acknowledgments: Financial support was provided by the Students Training in Advanced Research (STAR) Program through a UC Davis School of Veterinary Medicine Endowment Fund. Mentorship and oversight of this research, from proposal writing to data analysis, were provided by Drs. Beechler, Bird, Conrad and Kelly. Infrastructure and support were provided by Veterinary Wildlife Services in the KNP, with particular thanks to Dr. Peter Buss, Leana Rossouw and Guy Hausler for their tireless assistance in the field and laboratory. DNA sequencing equipment and expertise were provided by the Sharpton Lab at Oregon State University.
References:
- Belzer, C., et al. (2014). "Dynamics of the Microbiota in Response to Host Infection." PLOS ONE 9(7): e95534.
- de Vos, W. M. and E. A. de Vos (2012). "Role of the intestinal microbiome in health and disease: from correlation to causation." Nutrition reviews 70(suppl_1): S45-S56.
- Sharpton, T. J. and C. A. Gaulke (2015). "Modeling the context-dependent associations between the gut microbiome, its environment, and host health." MBio 6(5): e01367-01315.
- David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., ... & Biddinger, S. B. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature, 505(7484), 559.
- Ezenwa, V. O., Jolles, A. E., & O’Brien, M. P. (2009). A reliable body condition scoring technique for estimating condition in African buffalo. African Journal of Ecology, 47(4), 476-481.



